Large Language Models have shown promising results in their ability to encode general medical knowledge in standard medical questionanswering datasets. However, their potential application in clinical practice requires evaluation in domain-specific tasks, where benchmarks are largely missing. In this study semioLLM, we test the ability of state-of-the-art LLMs (GPT3.5, GPT-4, Mixtral 8x7B, and Qwen-72chat) to leverage their internal knowledge and reasoning for epilepsy diagnosis. Specifically, we obtain likelihood estimates linking unstructured text descriptions of seizures to seizure-generating brain regions, using an annotated clinical database containing 1269 entries. We evaluate the LLM’s performance, confidence, reasoning, and citation abilities in comparison to clinical evaluation. Models achieve above-chance classification performance with prompt engineering significantly improving their outcome, with some models achieving closeto-clinical performance and reasoning. However, our analyses also reveal significant pitfalls with several models being overly confident while showing poor performance, as well as exhibiting citation errors and hallucinations. In summary, our work provides the first extensive benchmark comparing current SOTA LLMs in the medical domain of epilepsy and highlights their ability to leverage unstructured texts from patients’ medical history to aid diagnostic processes in healthcare.
Introduction
Epilepsy is a chronic neurological disorder currently affecting 70 million people worldwide. It is characterized by a predisposition of the central nervous system to unpredictably generate seizures. About two thirds of patients suffer from focal epilepsies, which produce distinct seizure-related changes in sensation and behavior depending on the seizure onset zone (SOZ) in the brain. Patient reports on seizure symptoms - so-called seizure semiology - are therefore routinely recorded by clinicians and used as one crucial source of information for localization of the SOZ. This is particularly important for patients with drug-resistant epilepsies, for which surgical resection of the SOZ is the only potentially curable therapy option. For these patients, the clinicians’ task is to determine a confident and accurate estimate of the SOZ, as only then a recommendation for epilepsy surgery can be made.
In this direction, LLMs have sparked interest in their potential to leverage medical knowledge. However, there is a lack of systematic evaluation of LLMs’ understanding of specific clinical domains. Addressing this requires large-scale annotated text-datasets, systematic investigation of prompt designs, exploration of in-context learning strategies - all in comparison to problem evaluation technique followed by healthcare professionals in real world. In this work SemioLLM, we address several of these questions and show a first comprehensive investigation for benchmarking currently accessible LLMs in the domain of epilepsy.
Key Contributions and Findings
We highlight the contributions and key insights of this work as follows:
- First, we measure the correctness of the SOZ localization outputs generated by the LLMs. Second, we approximate the confidence in their outputs using entropy. Third, we perform a human evaluation to assess the LLMs' understanding, reasoning, and source retrieval abilities specific to the epilepsy domain.
- We estimate the impact of prompt strategies on the task employing five prompt styles: zero-shot, few-shot, Chain of Thought (CoT), few-shot CoT, and self-consistency (SC). Performance substantially increases with more sophisticated prompting strategies, highlighting the models' ability to leverage contextual information and domain-specific knowledge in epilepsy.
- With respect to model comparison, GPT-4 emerges as the top-performing model across all evaluation metrics. Mixtral8x7B, while competitive with GPT-4 in performance, exhibits tendencies to hallucinate in source citations and provides incomplete and partially incorrect reasoning. Notably, GPT-3.5 and Qwen-72B exhibit higher confidence levels in their outputs, albeit with reduced correctness.
Main Results
[A] Can LLMs give a 'correct' SOZ Localization Estimate?
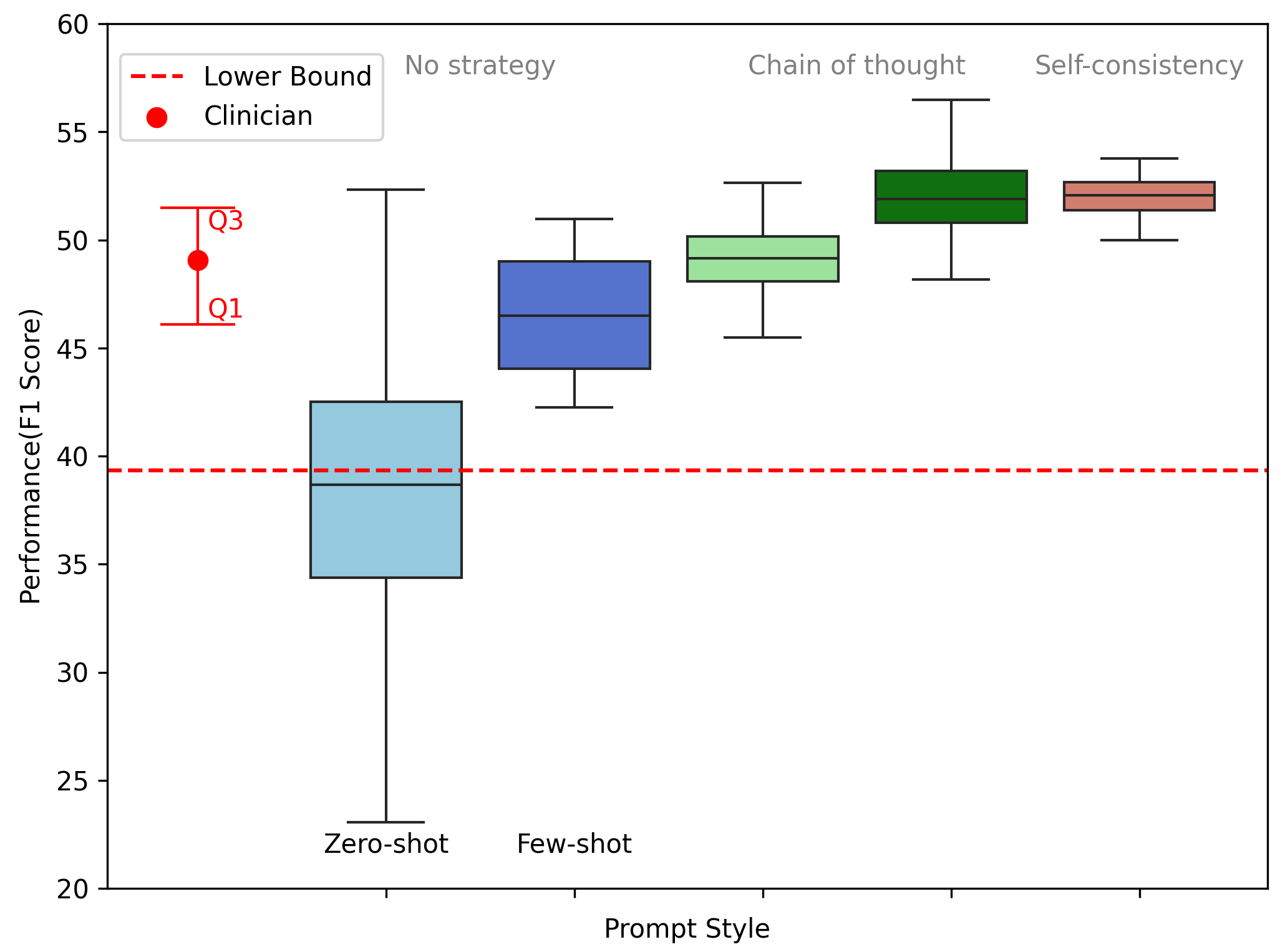
We report our localization results in Table below. For zero-shot prompting, only GPT4 shows significantly higher performance than the lower bound. With better prompting techniques, however, all models achieve substantially higher, and significantly better performance than expected by chance and is even comparable to clinical evaluation.
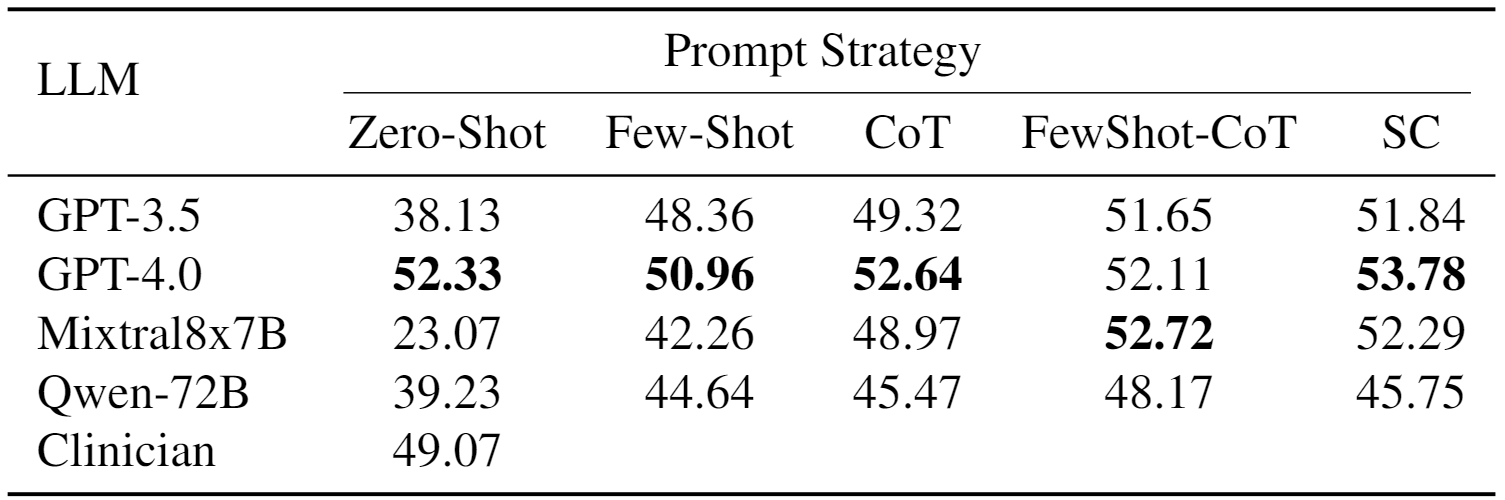
We also verify that the models get better in understanding the instruction and problem statement when introduced with more sophisticated prompting techniques.
[B] Are these models confident enough in their responses?
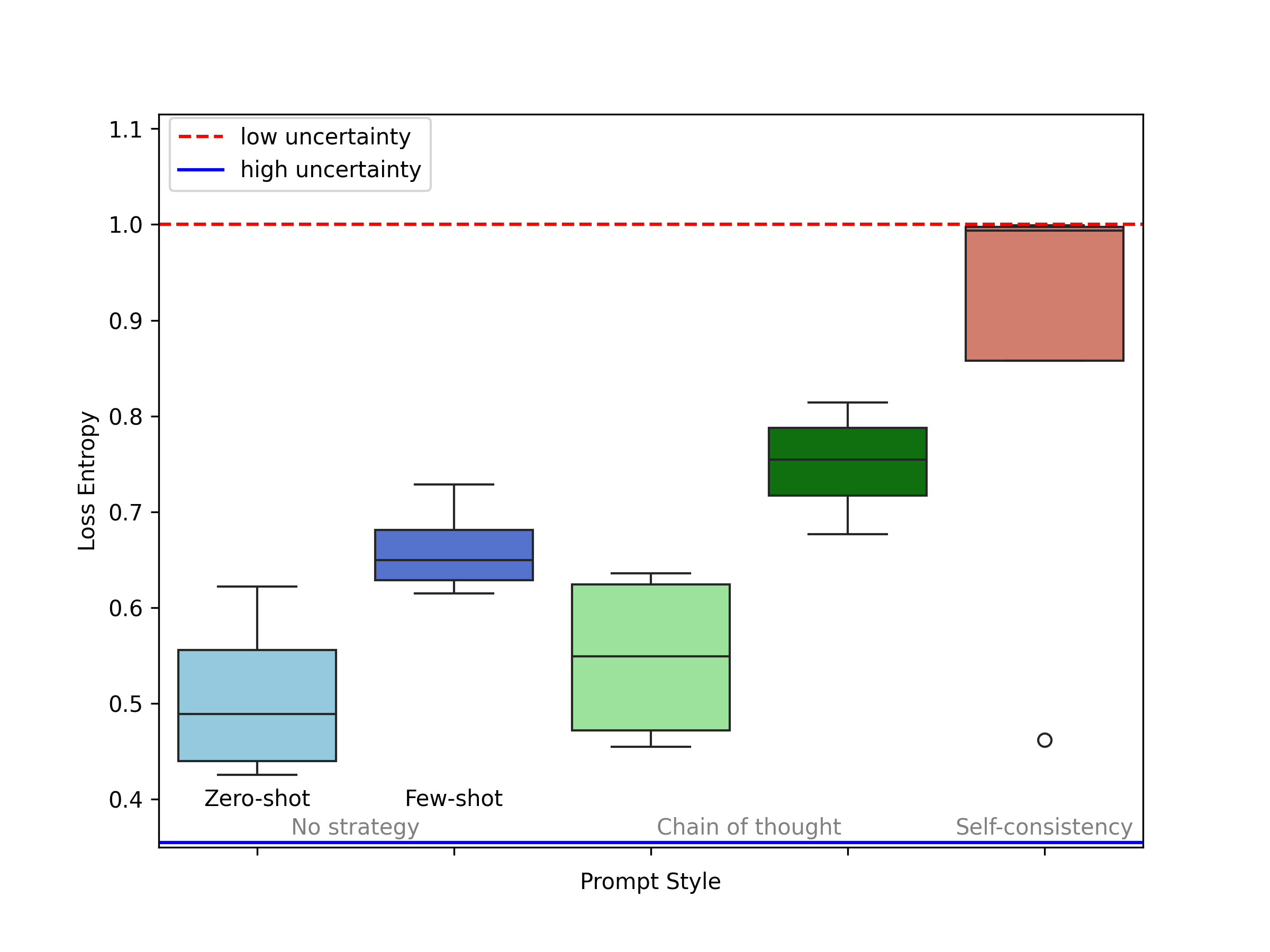
The likelihood output from the LLMs is more informative than a single class prediction, as it allows for understanding which classes the model considers plausible, and to what degree, rather than just which class it considers the ``winner''. We leverage this feature to approximate a confidence/uncertainty measure.
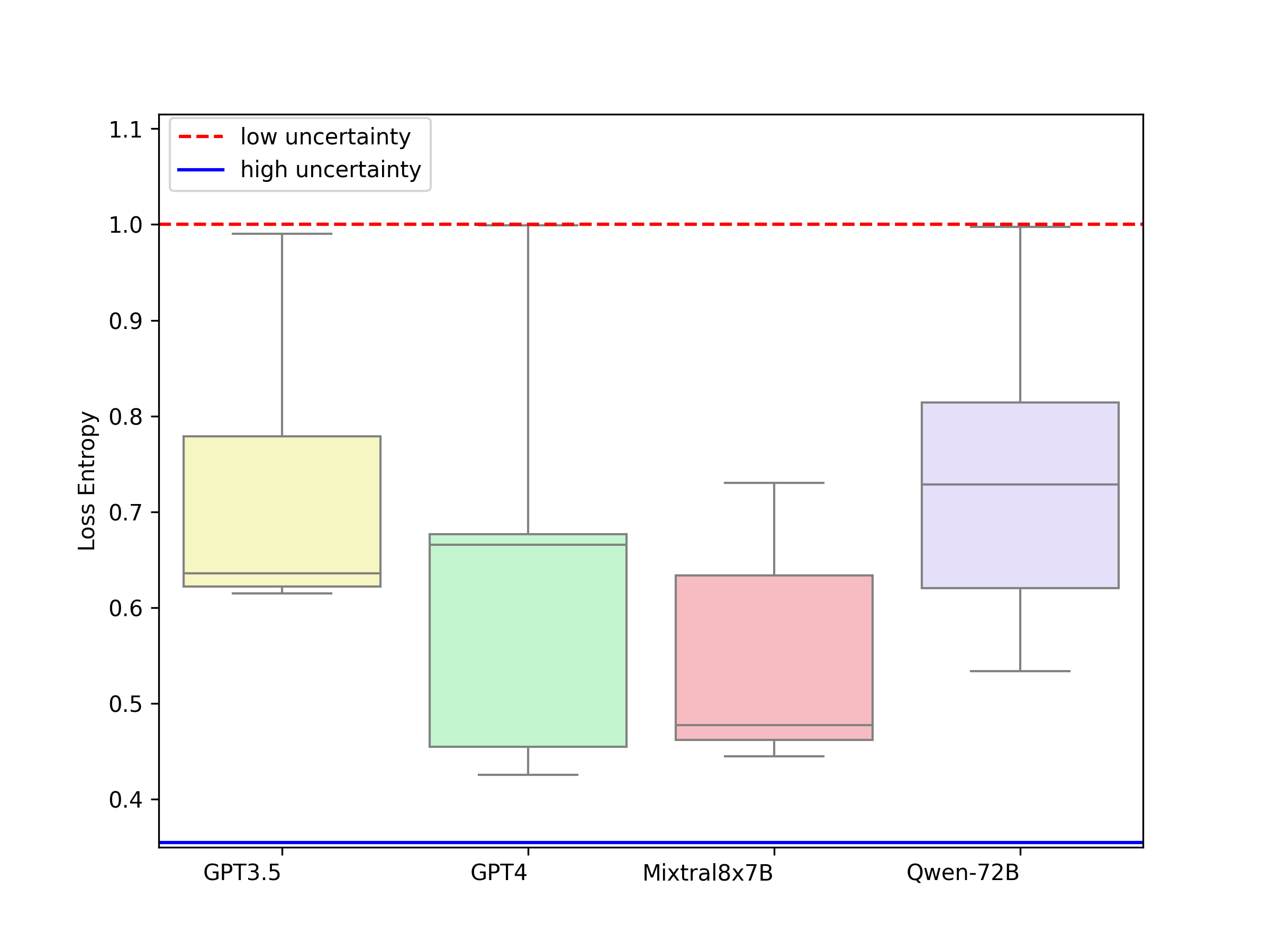
In the plot below, we can see a clear trend that the model becomes more confident when given some support examples in the case of Few-shot ICL (No strategy) and Few-Shot Chain of Thought. Evidently, it is least certain in zero-shot and most certain in self-consistency. We also compute model-wise entropy for all prompting techniques as shown in the figure in the right. It is crucial to note that GPT-3.5 and Qwen-72B did not perform well in terms of correctness evaluation but are much more confident about their responses. While the responses from the two best performing models GPT-4 and Mixtral8x7B are more grounded which makes them more trustworthy to be used in a clinical setting.
[C] Is there any clinical knowledge behind these responses?
In a rigorous clinical evaluation, we validate the reasoning, completeness, and source citation accuracy of our best-performing language models, GPT-4 and Mixtral8x7B. The task is to assess the correctness and completeness of the provided reasoning on a three-point scale (absolutely correct/complete, somewhat correct/complete, and not correct/complete).
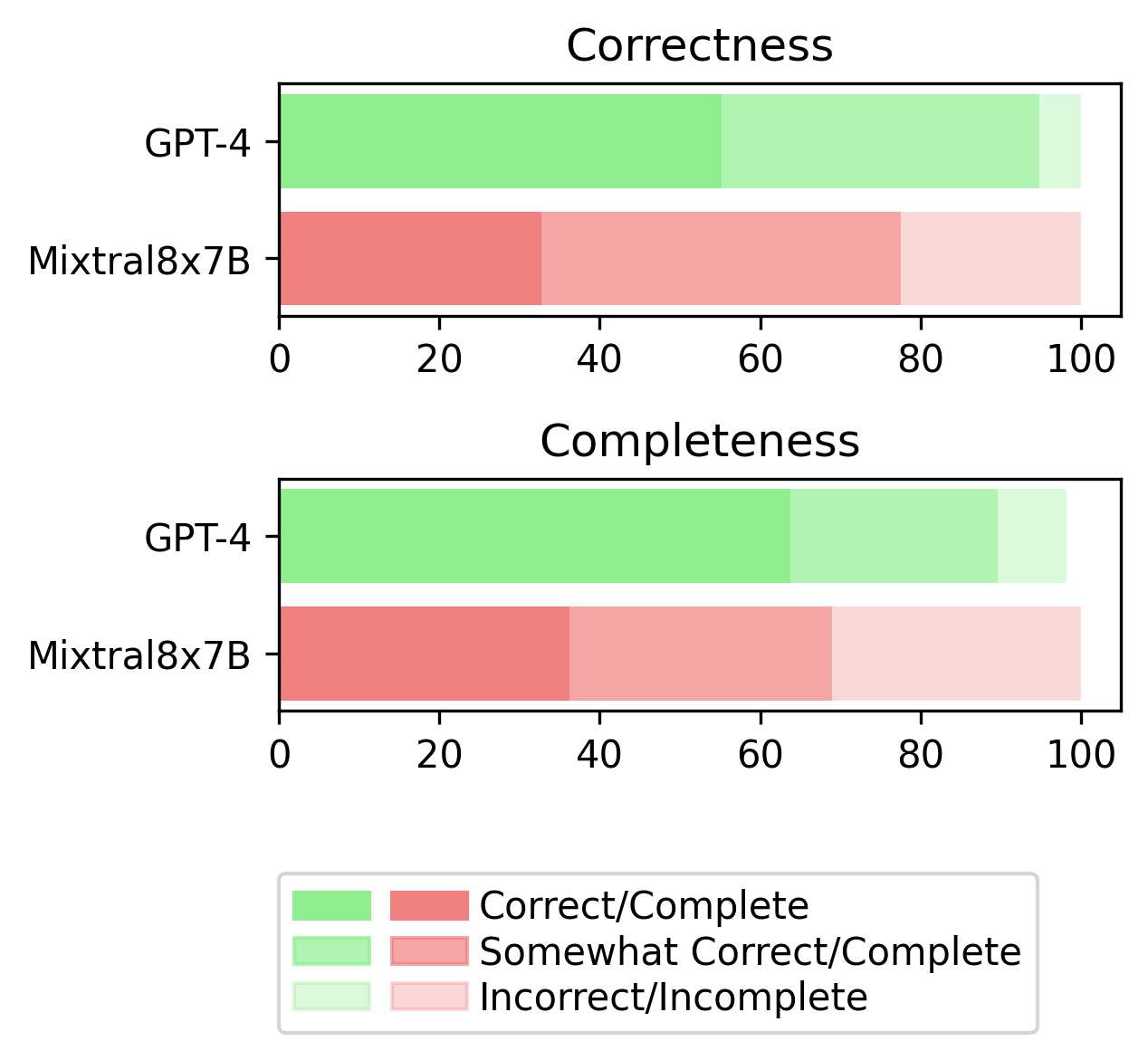
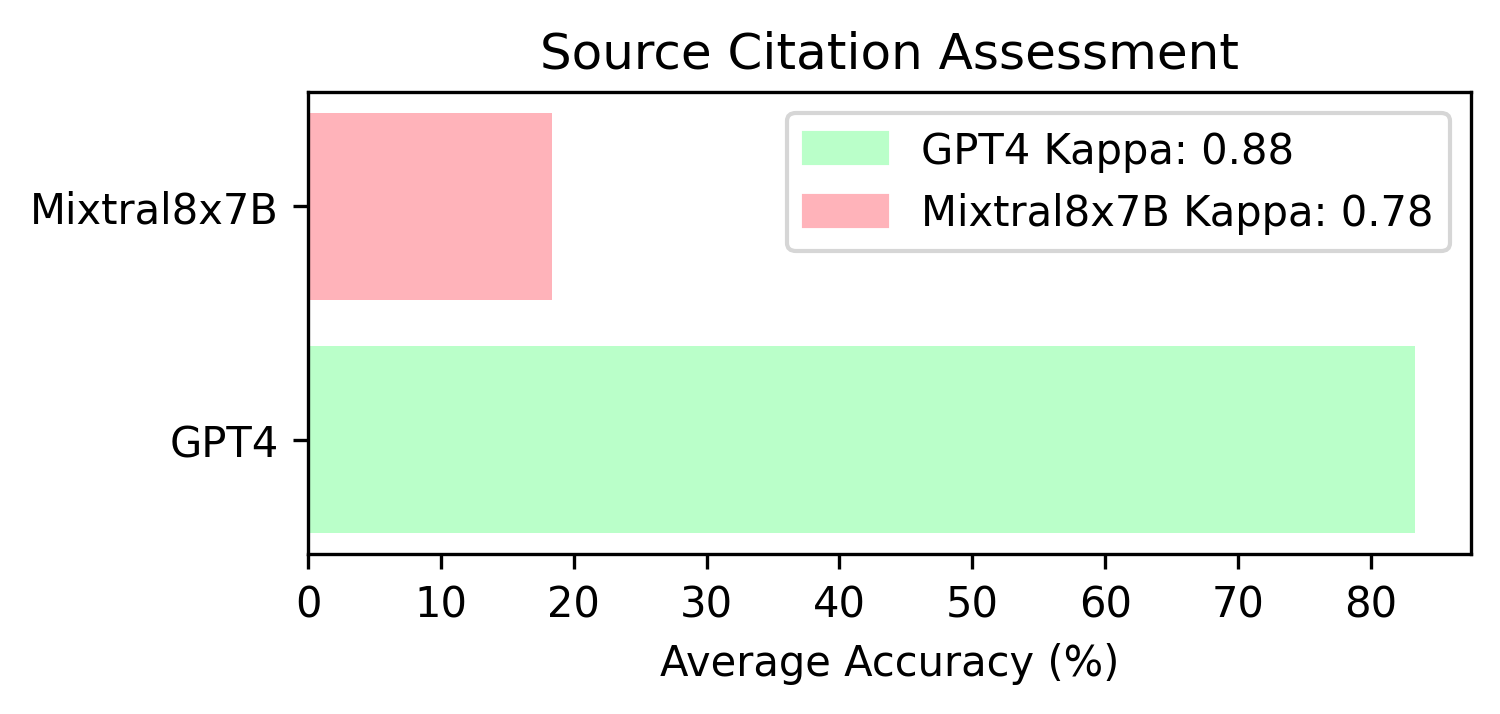
Additionally, we verify whether the sources cited by the models are correct or not. We only consider the source to be accurate if the author list, and exact title match verbatim.
Conclusion
In summary, our analyses provide evidence for the potential applicability of LLMs outside of the vastly used use case of standard medical QA datsets. In the domain of epilepsy, we show their potential contribution to improving the efficiency of diagnostic processes and treatment planning. Our approach can be extended to other domains, where unstructured text in medical history provides medical diagnostic information that can be verified using ground-truth datasets. Overall, our work contributes to the ongoing development and refinement of reliable and trustworthy language models for domain-specific applications, facilitating a comprehensive understanding of the models' performance and their potential for real-world use cases in the medical domain.